I am Rémy Vandaele, a new post-doc who started working for the DARE project this January 2020. I got a PhD degree in Computer Sciences from the University of Liège in Belgium. I am a researcher who works in the area of machine learning for computer vision. In this blog post, I will summarize the work I did during my PhD thesis, entitled « Machine Learning for Landmark Detection in Biomedical Applications ».

Figure 1 : 25 landmarks annotated on the image of a zebrafish larvae
The starting point of my thesis was about developing and comparing automated landmark detection methods. In this work, a landmark corresponds to a specific anatomic location on the image of a body (cf Fig. 1). It is defined by a type (tip of the nose, left corner of the left eye,…) and has coordinates. Detecting a landmark on a new image means finding its coordinates, given its type. In my work, I focused on landmark detection for two biomedical applications :
– Morphometry : biologists need to annotate landmarks on high resolution microscopy images of different types of bodies to understand the impact of their different experiments (e.g finding if the injection of a drug has impacted the size of a fish,…). Manual annotation is a long, tedious and error-prone process that limits the size of the experiments. Automation is severely needed in this area.
– CT-CBCT registration : in radiotherapy, a treatment plan (beam positioning & dosage) for all the sessions to come are computed on an initial, high resolution CT-scan. At each of the session, the patient is positioned on the table and needs to be positioned accordingly to its initial CT-scan. This is performed using a CBCT scan that is then registered to the CT-scan using the deformation computed between corresponding, manually annotated landmarks (see Fig. 2). Landmark positioning on 3D CT-scan is a slow process, that could be sped-up using automation.
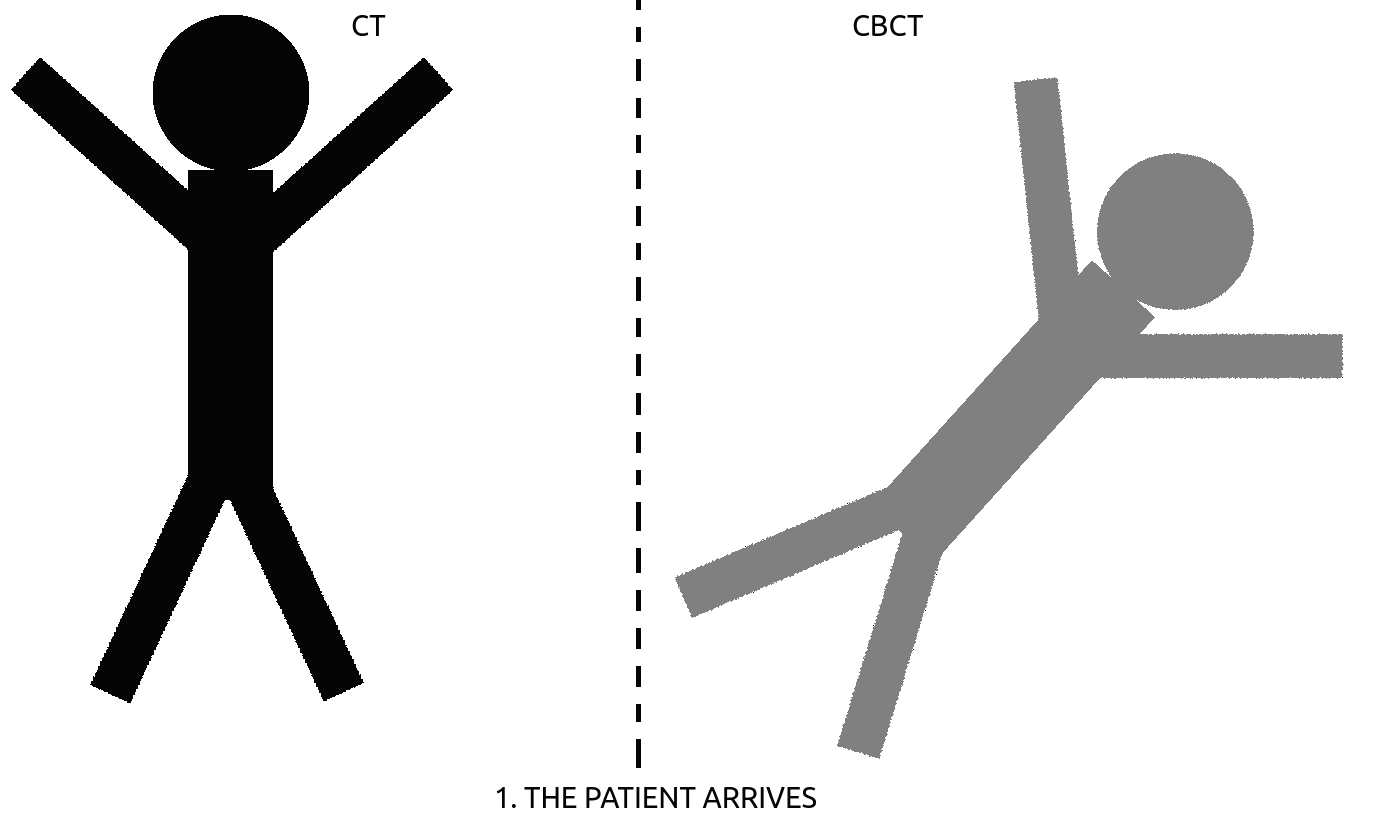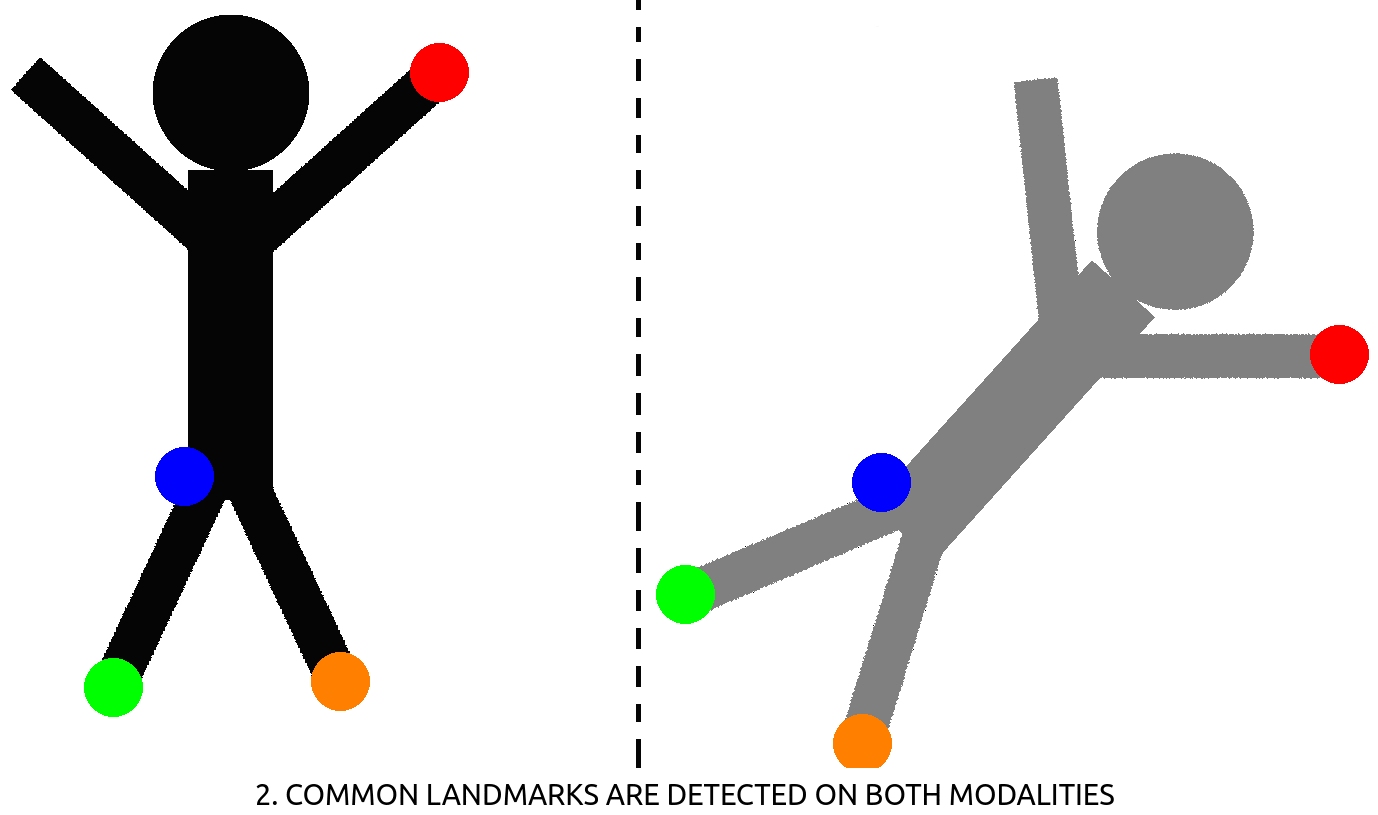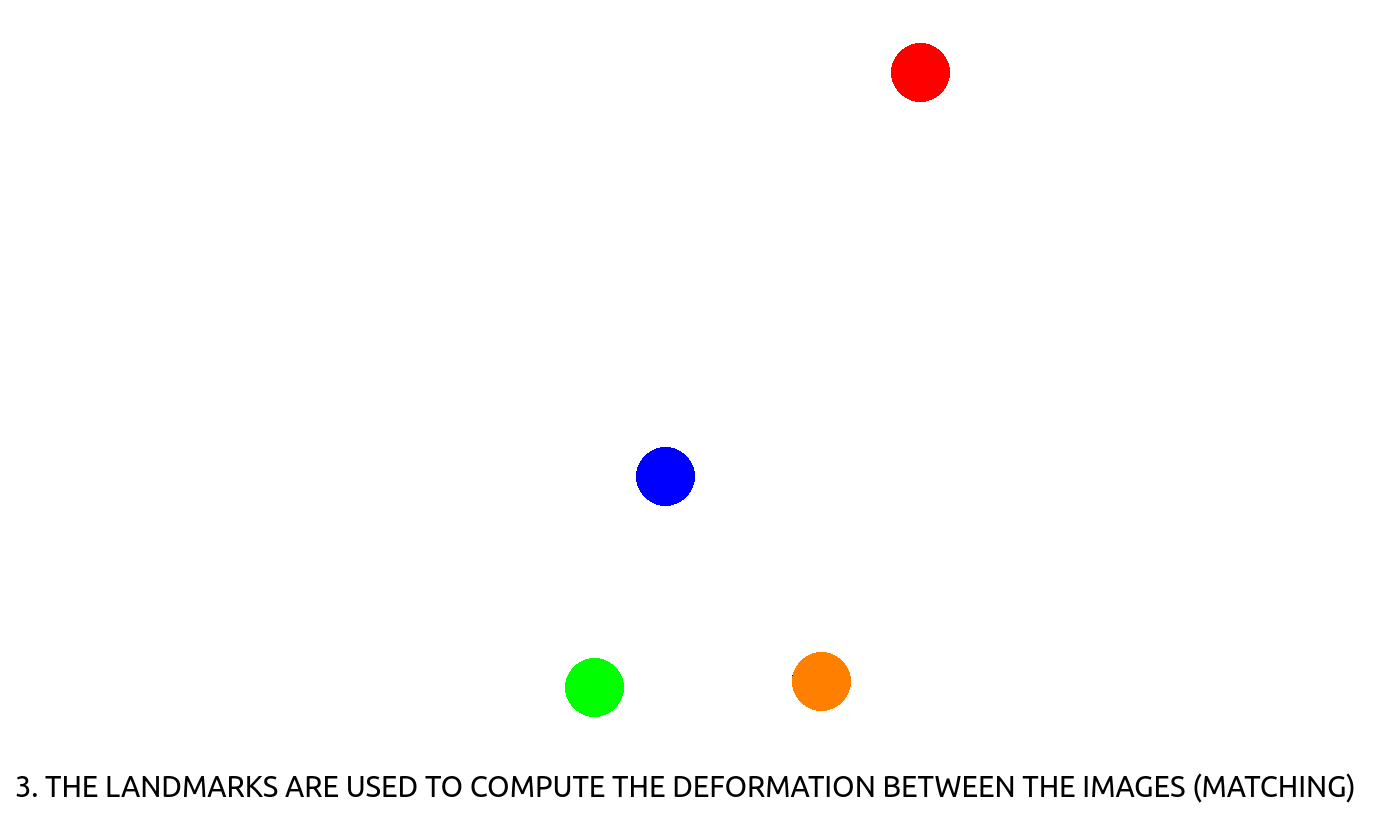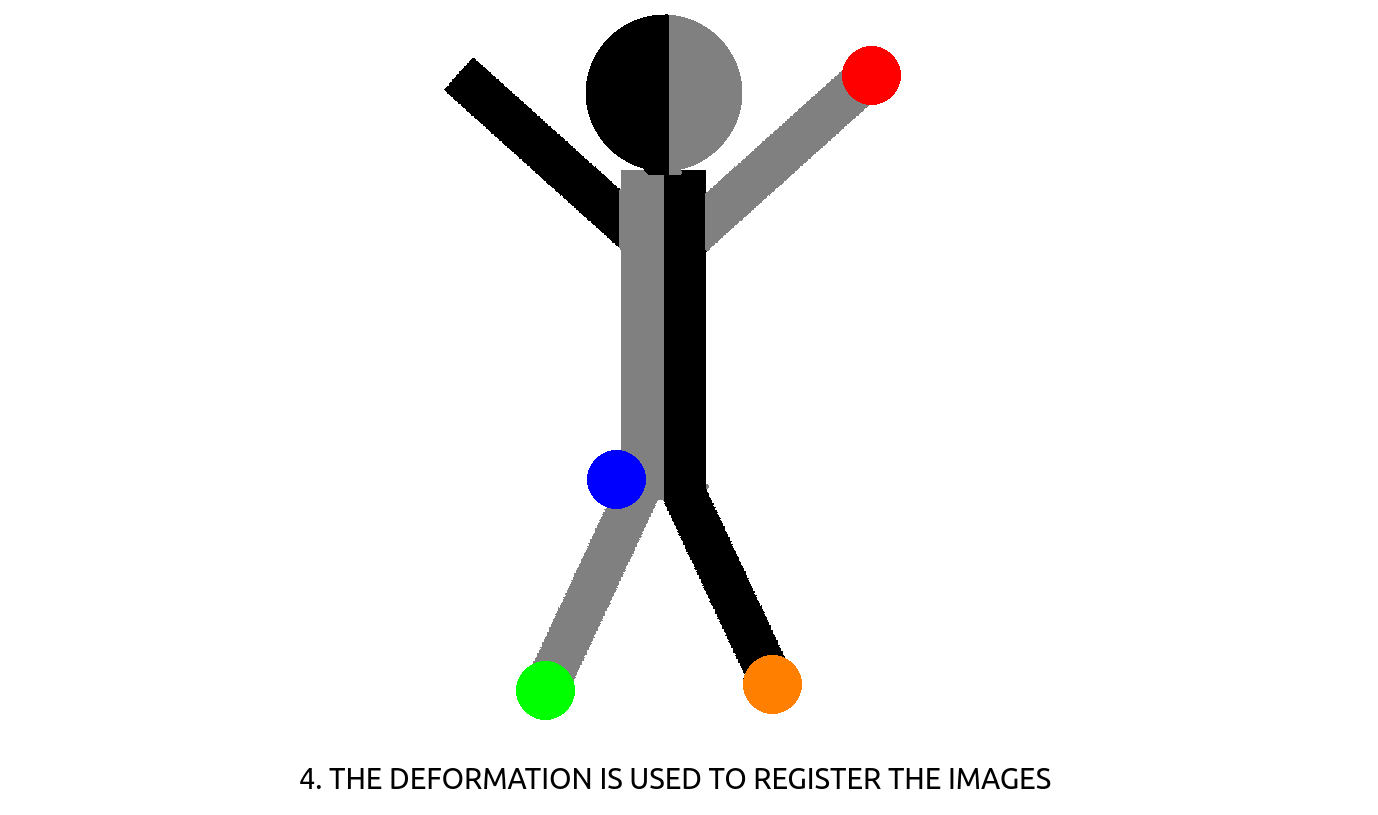
Figure 2: How to register images using landmark detection
My work on this topic can be divided into three parts :
- I developed a 2D method based on binary image patch classification using a Random Forest [1] algorithm. During this work [2],[3],[4] , I was able to show (1) the relevance to use multi-resolution patch descriptors in order to increase the accuracy of the landmark detection algorithm (2) we could reduce the search zone of the landmark, and thus greatly increase the speed of detection, without losing accuracy. Those algorithms were implemented into an open-source client-server application for the analysis and sharing of large (bio)images, Cytomine [5] [6].
- I expanded this method to 3D in order to show that we could use my method for 3D CT to CBCT image registration [7].
- I studied the impact of state-of-the-art post-processing methods [8] (methods taking into account the relative position between the detected landmarks to refine their locations) in the context of biomedical applications, where the number of images and landmarks is significantly lower than in typical face detection applications where landmark detection is widely used. In this context, I showed that those methods brought little to no increase in detection accuracy, and proposed a new post-processing method suited for landmark detection in biomedical applications.
Future perspectives regarding this work will consider deep learning classification and regression models in combination with data augmentation approaches. This research is currently pursued at the University of Liège.
References
[1] Geurts, Pierre, et al. “Extremely randomized trees.” Machine learning 63.1 (2006): 3-42.
[2] Vandaele, Rémy, et al. “Automatic cephalometric x-ray landmark detection challenge 2014: A tree-based algorithm.” ISBI (2014).
[3] Wang, Ching-Wei, et al. “Evaluation and comparison of anatomical landmark detection methods for cephalometric x-ray images: a grand challenge.” IEEE transactions on medical imaging 34.9 (2015): 1890-1900.
[4] Vandaele, Rémy, et al. “Landmark detection in 2D bioimages for geometric morphometrics: a multi-resolution tree-based approach.” Scientific reports 8.1 (2018): 1-13.
[5] Marée, Raphaël, et al. “Collaborative analysis of multi-gigapixel imaging data using Cytomine.” Bioinformatics 32.9 (2016): 1395-1401.
[6] Rubens, Ulysse, et al. “BIAFLOWS: A collaborative framework to benchmark bioimage analysis workflows.” bioRxiv (2019): 707489.
[7] Vandaele, Rémy, et al. “Automated landmark detection for rigid registration between the simulation CT and the treatment CBCT.” Acta Stereologica (2015).
[8] Vandaele, Rémy. “Machine Learning for Landmark Detection in Biomedical Applications.” Diss. Université de Liège, Liège, Belgique, 2018.
