Improving future projections of changes in climate and the terrestrial biosphere by exploiting the power of the past is the main aim of SPECIAL (Sandy’s Palaeo Environments and Climate Analysis Group) research group at the University of Reading. These reconstructions provide insights into how vegetation responds to climate changes and human activities. Creating reconstructions of vegetation patterns at the regional and continental scale has widely relied on biomisation, a technique of allocating pollen taxa to a range of plant functional types (PFTs) and how these PFTs combine to form biomes. The biggest benefit of this method is that it is simple, and allocations can be optimized with relatively small pollen datasets. The biggest drawback is that it requires expert knowledge of vegetation to make these allocations, and this cannot straightforward handle the variability of expressions of a biome.
A new paper, led by SPECIAL’s PhD candidate Esmeralda Cruz-Silva, outlines a new method to quantifiably reconstruct past vegetation patterns with pollen that does not rely on expert judgement and takes into account the within-biome variability:
A new method based on surface-sample pollen data for reconstructing palaeovegetation patterns. Journal of Biogeography
Biomisation
Before we get into the new model, let’s recap on biomisation, the benefits of it and its limitations. Back in 1996, Prentice et al.,developed the method called biomisation. The principle of biomisation method is to assign pollen taxa to PFTs based on leaf form (e.g., broad-leaved/ needle-leaved), stature (e.g., tree/shrub), leaf type, phenology (e.g. evergreen/deciduous) and climate tolerance. Taxa within a PFT are assumed to have similar responses to physical and biotic environmental factors. Major vegetation types or biomes are then characterised as assemblages of PFTs, where the PFTs included represent either the dominant taxa or taxa that are considered characteristic of the biome. This method is relatively simple, can be optimized with small pollen sample sizes and it has been shown to produce robust reconstructions of vegetation patterns. However, there are some limitations:
- The allocation of pollen into PFTs and biomes is done initially through expert knowledge of vegetation, but this cannot easily prevent that there is no direct relationship between pollen abundance and abundance of plants, meaning some plants are either under-represented or over-represented in a biome, nor easily capture the within-variability of plants within each biome and gradients that can be seen from the northern to southern limits of a biome.
- Some biomes are characterized by a subset of the PFTs present in another biome, which can cause in certain cases, sudden unreal changes of biomes between samples that are together.
- So far, Biomisation has not been addressed to identify palaeovegetation communities that have no comparison to modern vegetation types because whilst the species might be the same, their combinations are different to historic communities. We refer to these as non-analogue vegetation types.
Whilst this method has been played a key role to date in reconstructing past vegetation patterns, there is a need to fine-tune this process and make it more quantifiable with less reliance on expert judgement.
New dissimilarity-based technique
This new method, a collaboration between UoR’s SPECIAL team and Colin Prentice, have devised a way to overcome the standard biomisation limitations. It quantifies the likelihood that a sample belongs to a given biome and allows discrimination of non-analogue vegetation types. The method involves calculating a dissimilarity index (which takes account of the within-biome variability) between a given pollen sample and every biome, and thereby assessed the likelihood that a sample belongs to a particular biome.
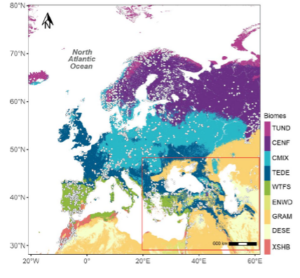
As a test, this method was used to reconstruct modern vegetation of the Eastern Mediterranean-Black Sea Caspian Corridor (EMBSeCBIO) region. The EMBSeCBIO region is characterized by strong temperature and precipitation gradients and topographic heterogeneity which has resulted in clear patterns in biome distribution within a relatively limited geographic space. The results from this new approach were then compared to the reconstructions made using the standard biomisation technique.
Modern pollen samples (derived from the SPECIAL Modern Pollen Data Set, SMPDS), were assigned to biomes based on potential natural vegetation data and were used to characterize biomes according to the within-biome means and standard deviations of the abundances of each taxon. These were used to calculate a dissimilarity index between any given pollen sample and every biome, and thus assign a pollen sample to the most likely biome. A threshold value was calculated for each biome which identifies samples that fall outside the acceptable range of likelihoods for biome assignment and hence can be used to distinguish non-analogue vegetation.
The workflow for the process is summarised in Figure 2 and you can read the full methodology in the paper.

The results from this study demonstrate that there is considerable within-biome variability in taxon abundance with changes in individual taxa along climate and environmental gradients. Whilst the most abundant taxa might be the same in both methodologies, biomisation does not fully grasp this detail of the importance of less dominant vegetation types and how even dominant types can change in composition between warm and cold ends of a biome. The balanced accuracy obtained for the EMBSeCBIO region using the new method was better than that obtained using biomisation (77% versus 65%). When the method was applied to high- resolution fossil records, 70% of the evaluated entities showed more temporally stable biome assignments than obtained with the biomisation method. The technique also identifies likely non- analogue assemblages in a synthetic modern data set and in fossil records.
Whilst this method requires an extensive modern pollen dataset, more than required for biomisation, it is considerably less data-demanding that some other approaches such as the Landscape Reconstruction Algorithm (LRA). Despite the larger dataset required, the actual concept is simple and omits the need for allocation by experts into PFTs and biomes and it accounts for the within-biome variability in taxon abundances. Another benefit of this approach is that under- or-over representation of some species in pollen assemblages are circumvented. Even poorly sampled biomes, such as deserts, were accurately reconstructed, something that the traditional biomisation was not able to do well. This new method provides a more accurate and stable reconstruction of vegetation than the biomisation method by creating a more nuanced way of examining past vegetation changes and transitions.
Cruz-Silva, E., Harrison, S.P., Marinova, E. & Prentice, I.C. 2022. A new method based on surface-sample pollen data for reconstructing palaeovegetation patterns. Journal of Biogeography.
 Esmeralda is a PhD candidate in the SPECIAL team (Sandy’s Palaeo Environments and Climate Analysis) at the University of Reading. Her thesis is on the interactions of climate, vegetation and human activities in the circum-Mediterranean region.
Esmeralda is a PhD candidate in the SPECIAL team (Sandy’s Palaeo Environments and Climate Analysis) at the University of Reading. Her thesis is on the interactions of climate, vegetation and human activities in the circum-Mediterranean region.
This project was funded by the ERC project GC 2.0 (Global Change 2.0: Unlocking the past for a clearer future, grant number 694481) and also by Colin’s ERC funded REALM project.