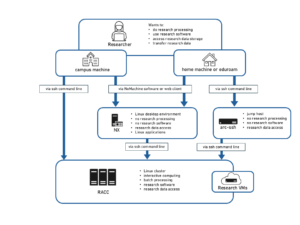
Knowledge Base
- /
- /
- /
 Using R and R Studio on RACC
Using R and R Studio on RACC
The “parallel” library is used for parallel processing!
Uses an internal R dataset called MASS which has ‘Boston’ and ‘iris’ data.
This can be used in a batch script on the RACC.
'R script for parallelising.
library(parallel)
library(MASS)
starts <- rep(100, 40)
fx <- function(nstart) kmeans(Boston, 4, nstart=nstart)
numCores <- detectCores()
numCores
system.time(
results <- lapply(starts, fx)
)
system.time(
results <- mclapply(starts, fx, mc.cores = numCores)
)
x <- iris[which(iris[,5] != "setosa"), c(1,5)]
trials <- seq(1, 10000)
boot_fx <- function(trial) {
ind <- sample(100, 100, replace=TRUE)
result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit))
r <- coefficients(result1)
res <- rbind(data.frame(), r)
}
system.time({
results <- mclapply(trials, boot_fx, mc.cores = numCores)
})
Output shows the processing speeds using the different libraries lapply and mclapply