By Alastair Culham
Welcome to #AdventBotany 2019 and the start of another journey into quirky, curious, historical and, above all, botanical information about the plants associated with the winter season.
This year I’m expanding on John Warren’s story of Tangerines and virgin birth told in 2015 with my explorations into the identity of the smaller fruited citrus. I’ve often seen and bought fruit variously under the names Clementine, Satsuma and Tangerine and felt that, to an extent, they were all rather similar. However I am not the only one to have wondered about the mysteries of Citrus.
There are four species of Citrus widely treated as the wild ancestors of most cultivated ones: C. reticulata, C. maxima, C. medica, and C. micrantha.
All cultivated Citrus are eastern in origin including the wild mandarin.

Ideas of the derivation of cultivated Citrus from these four ancestral species is depiceted in an excellent illustration from Recent insights on Citrus diversity and phylogeny
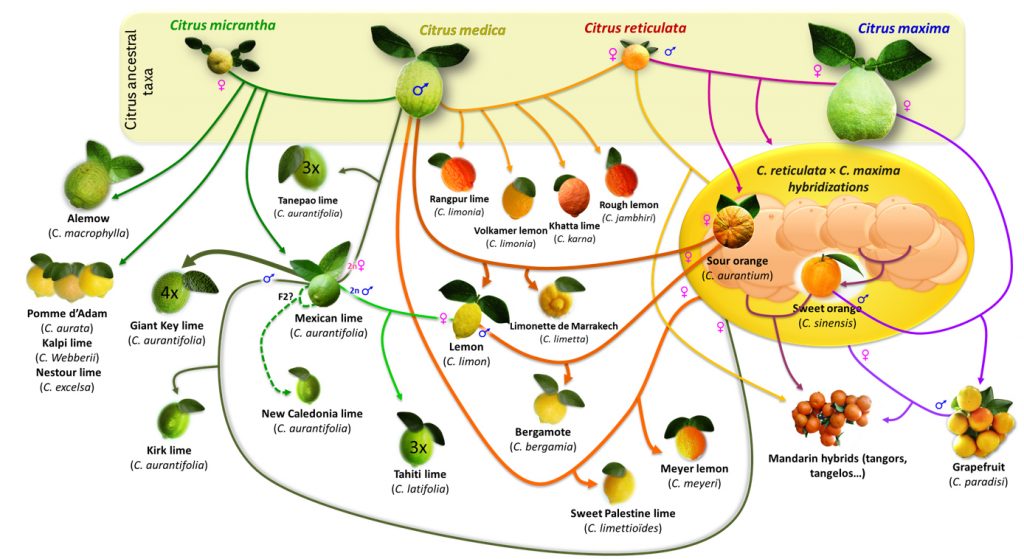
Genomic studies of Citrus have shown more detail on the relationships of genomes in the cultivated species (Curk et al., 2014; Velasco & Licciardello 2014; Wang et al. 2017) including maps of the nine basic citrus chromosomes and have identified candidate genes for asexual seed production (apomixis via nucellar polyembryony) which has been important in the domestication of these fruit. In 2018 (Wu et al. 2018) further insights into the genomics of Citrus investigated ten wild species, using genomic, phylogenetic and biogeographic analysis. They propose that “citrus diversified during the late Miocene epoch through a rapid southeast Asian radiation that correlates with a marked weakening of the monsoons. A second radiation enabled by migration across the Wallace line gave rise to the Australian limes in the early Pliocene epoch.”Their research suggests that sweet orange and mandarins are extensively interrealted and have evidence of pummelo genes through hybridization. Work by Oueslati et al. (2016) offers an insight into quite how closely related the various Citrus cultivars are
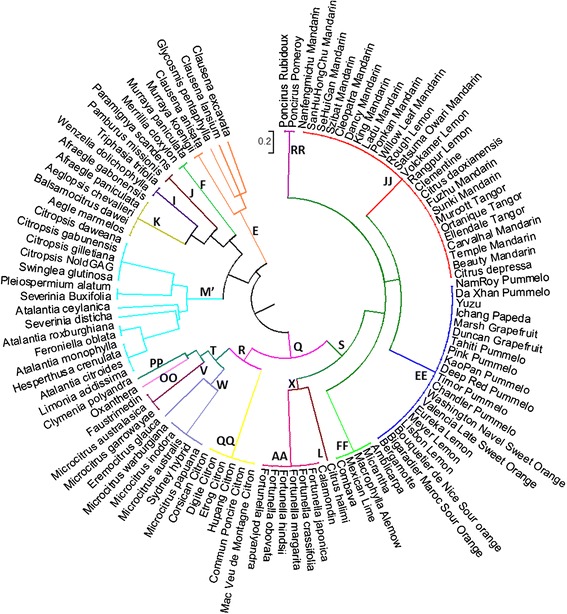
Published online 2016 Aug 18. doi: 10.1186/s12863-016-0426-x]

The article in independent.ie is perhaps the most accessible suggesting:
Clementines are the sweetest of the small orange citrus, and have a skin that peels fairly easily due to the pith thickness.
Tangerines gain their name from the north African exports of Citrus via the port of Tangiers. Tangerines are a form of mandarin orange and are the hardest of these three to peel (you can see the thin peel with only a small albedo) but have a richer, sweeter flavour than the others.
Satsumas have an easy to peel skin due to a thick but loose albedo (the white layer under the orange skin) so the central segments can be freed readily from the peel.
So I can now tell what to expect when shopping for these winter season fruit.
References
Curk F, Ancillo G, Ollitrault F, Perrier X, Jacquemoud-Collet J-P, Garcia-Lor A, et al. (2015) Nuclear Species-Diagnostic SNP Markers Mined from 454 Amplicon Sequencing Reveal Admixture Genomic Structure of Modern Citrus Varieties. PLoS ONE 10(5): e0125628. https://doi.org/10.1371/journal.pone.0125628
Luro, F., Curk, F. Froelicher, Y. & Ollitrault, P., Recent insights on Citrus diversity and phylogeny. Publications du Centre Jean Bérard, (2017) https://books.openedition.org/pcjb/2169
Oueslati, A., Ollitrault, F., Baraket, G. et al. Towards a molecular taxonomic key of the Aurantioideae subfamily using chloroplastic SNP diagnostic markers of the main clades genotyped by competitive allele-specific PCR. BMC Genet 17, 118 (2016) doi:10.1186/s12863-016-0426-x
Velasco, R., Licciardello, C. A genealogy of the citrus family. Nat Biotechnol 32, 640–642 (2014) doi:10.1038/nbt.2954 [Full text]
Wang, X., Xu, Y., Zhang, S. et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet 49, 765–772 (2017) doi:10.1038/ng.3839
Wu, G., Prochnik, S., Jenkins, J. et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 32, 656–662 (2014) doi:10.1038/nbt.2906 [Preprint here]
Wu, G., Terol, J., Ibanez, V. et al. Genomics of the origin and evolution of Citrus. Nature 554, 311–316 (2018) doi:10.1038/nature25447



